Kirsten Krause
Job description
Jeg ønsker å forstå hvordan planter tilpasser seg forskjellige omgivelser og utfordringer. Mitt tidligere arbeid på cellulære nettverker i modellplanten Arabidopsis thaliana har ledet meg de siste årene til å utforske snylteplanteslekten Cuscuta som lever av de næringsstoffer andre planter produserer. De molekylære mekanismer som parasittene bruker å tilpasse seg verten og kamuflere seg selv slik at infeksjonen ikke oppdages av vertsplanten kan ha bioteknologisk verdi og er derfor blant de spørsmålene jeg håper å få løst i min karriere.
I min rolle som leder av Forskerskolen "Photosyntech" (2022-2030), en nasjonal utdanningsplattform for stipendiater som arbeider innenfor alle disipliner av plante- og algevitenskap, håper jeg å kunne videreformidle min lidenskap for forskning og innovasjon til neste generasjon av forskere.
The 50 latest publications is shown on this page. See all publications in Cristin here →
Research interests
Functional Parasitic Plant Genomics:
Parasitic plant/host plant interactions
 Parasitic plants live as parasites on and from other plants. This creates huge losses in agriculture when a field of crop plants is infected. Knowledge on the biology of these plants is therefore of high importance to combat them, but also to make use of their unusual traits.
Parasitic plants live as parasites on and from other plants. This creates huge losses in agriculture when a field of crop plants is infected. Knowledge on the biology of these plants is therefore of high importance to combat them, but also to make use of their unusual traits.
We have over the last years developed a set of tools for the parasitic plant genus Cuscuta spp. (dodder) that allow us to approach the molecular mechanisms underlying host plant/parasitic plant interactions. These include a sequenced genome (the first parasitic plant genome sequence), gene expression profiles for different species, different developmental stages and different growth conditions, proteomic data, sterile callus and shoot cultures, protocols for transient transfections of cells and an extensive microscpic image database including time lapse videos.
Selected publications:
- Fischer K et al.: Mol. Phylogenet. Evol., DOI: 10.1016/j.ympev.2024.108243, 2024.
- Bawin T et al.: Plant Phys., https://doi.org/10.1093/plphys/kiad505, 2023.
- Zangishei Z. et al.: Plant Phys., https://doi.org/10.1093/plphys/kiac331, 2022.
- Fischer K. et al.: Front. Plant Sci. 12, 641924, 2021.
- Förste F. et al.: Physiol. Plant. 168, 934-947, 2020.
- Olsen S & Krause K: Plant Meth. 15, 88, 2019.
- Vogel A et al.: Nature Commun. 9, 2515, 2018.
Cell wall degrading enzymes from parasitic plants
![]() Parasitic plants use hydrolytic enzymes to break down cell walls of their host plants but are able to protect their own cell walls from enzymatic attack.
Parasitic plants use hydrolytic enzymes to break down cell walls of their host plants but are able to protect their own cell walls from enzymatic attack.
We study the difference between cell wall composition in the parasite Cuscuta and its hosts and analyze the enzymes involved. This knowledge can help to develop novel, more specific enzyme cocktails for plant biomass hydrolysis. In addition, it helps us understand why some plants are not susceptible to the parasite.
Selected publications:
- Olsen S. et al.: Journal of Experimental Botany 67, 695-708, 2016.
- Johnsen H.R., et al.: New Phytologist 207, 805-816, 2015.
-Johnsen H.R. & Krause K: International Journal of Molecular Sciences 15, 2014.
Functional Plant Genomics (FunGen): Arabidopsis thaliana
Integration of plastids into the regulatory network of plant cells
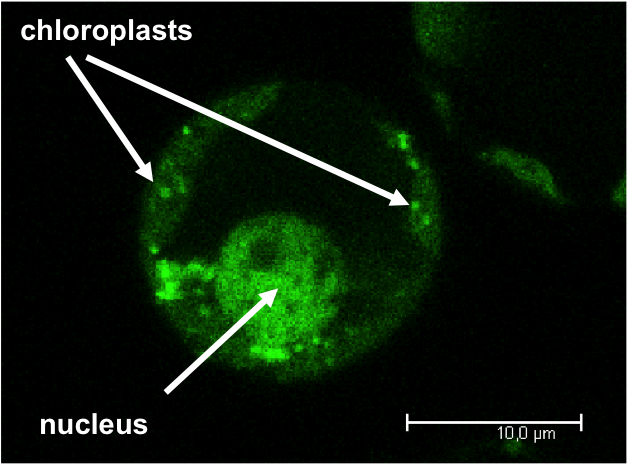 The genetic information in plant cells is distributed between three compartments (nucleus, plastids and mitochondria). To function properly, these genomes are need to communicate with each other to synchronize their expression.
The genetic information in plant cells is distributed between three compartments (nucleus, plastids and mitochondria). To function properly, these genomes are need to communicate with each other to synchronize their expression.
We are investigating how DNA binding proteins that are targeted within the cell with a special focus on proteins that have functions in the nucleus and in the chloroplasts. We are further interested how this mutual communication between the plant genomes contributes to a fine-tuning and with it a harmonized expression of the genomes.
Selected publications:
- Teubner, M. et al.: Plants 9, 367, 2020.
- Teubner, M. et al.: Plant J. 89, 472-485, 2017.
- Fuss, J. et al.: New Phytologist 200, 1022-1033, 2013.
- Krause, K. & Krupinska, K.: Trends Plant Sci 14, 194-199, 2009.
Teaching
- Bio-2009 Green Biotechnology and Bioenergy
- Bio-3118 Microscopical Imaging
- Bio-3005/Bio8005 "MicroPlants" Seminar
Member of research group
CV
1994 Diploma in Biology from the University of Hamburg, Germany.
1995-2000 PhD studies at the Universities of Cologne and Kiel.
2000-2002 Postdoc at the University of Arizona, USA, with a grant from the Geman Academic Exchange Organization (DAAD).
2002-2006 Research Assisant at the University of Kiel, Germany. Venia legendi 2006.
2006-2012 Researcher and YFF-Professor at the Department of Biology, University of Tromsø, Norway.
Since 2012 Permanent position as Professor for Plant Molecular Biology at the Department for Arctic and Marine Biology, UiT - Campus Tromsø, Norway.
2010 - 2018 Leader of the "Microorganisms and Plants Research Group" at the Department for Arctic and Marine Biology, UiT - Campus Tromsø, Norway.
2016 - 2021 Holder of a ToppForsk Grant from Tromsø Research Foundation
2017 - 2025 Member of the Arctic Centre for Sustainable Energy (ARC)
2022 - Leader of a National Graduate School, PhotosynTech
2022 - Faculty Board Member
2022 - Vice president (until 2024) and president (since 2024) of the Scandinavian Plant Physiology Society (SPPS)